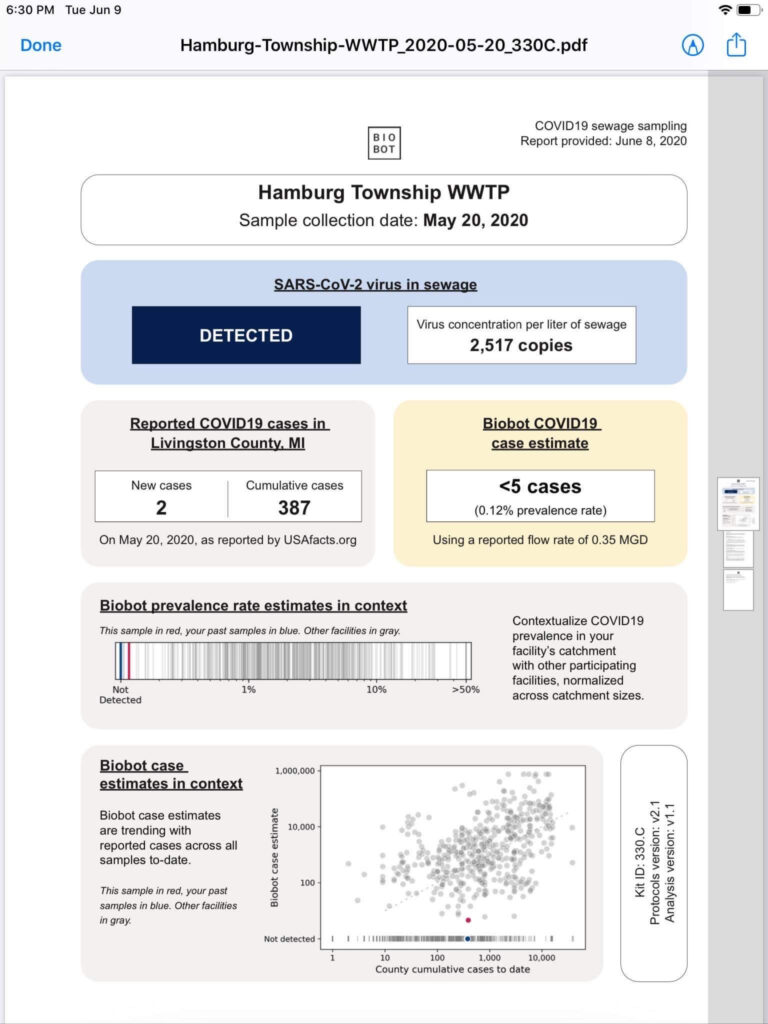
Covid-19 is currently a hot topic, environmental health and engineering is no exception. Wastewater is now in international news because of it! This Reuters article from 19 June 2020 for example shows that researchers found RNA from Covid-19 in Milan and Turin’s wastewater in December 2019 before China reported the first cases on 31 December 2019! The Italian National Institutes of Health examined 40 sewage samples collected in northern Italy between October 2019 and February 2020 and found that samples in Milan and Turin from 18 December 2019 showed SARS-Cov-2. Monitoring sewage for health purposes is known as “wastewater-based epidemiology” (WBE).
Early WBE
Using wastewater to track populations is not a new idea. It was first proposed by Christian Daughton in 2001 to track illicit drug use. You can read his paper here. As a former wastewater teacher of mine, COL Timmes, liked to say: “everyone passes through us.” Generally, he meant that you can’t easily hide from the central sewage system. In more polite terms raw wastewater is a reservoir of excretion products such as: parent compounds, metabolites, and genetic material. The earliest widespread use of WBE (then called “sewage epidemiology”) was in 2005 to monitor for illicit drugs which you can find here. After this early case WBE gained traction. At least Australia, Belgium, Germany, Ireland, Italy, the Netherlands, Norway, Spain, South Korea, the United Kingdom, and the United States use WBE to monitor illicit drug use. After this initial use WBE started to take off in public health circles and WBE started to be used to track broader chemical public health indicators, for instance alcohol consumption in Norway, counterfeit medicine distribution in the Netherlands, and even tobacco use in Italy.
Environmental engineers and public health officials eventually realized that any excreted substance that has known kinetic pathways in wastewater could be used to reverse engineer the initial concentration. All these early methods focused on chemicals and were based around mass spectrometry. WBE was then and is still used to study exposure to chemicals or pollutants such as pesticides, herbicides, and flame retardants. After the sewer’s viability as a surveillance network was established, someone around 2008 realized with some work they could use quantitative polymerase chain reaction methods (qPCR) to amplify, detect, and quantify genetic material.
WBE basics
WBE’s popularity continues to increase because exclusive reliance on testing of individuals is slow, costly, and generally impractical. WBE also often serves as a disease early warning indicator because asymptomatic or prodromal individuals typically don’t get tested and there may be underdiagnosis. In cases like this WBE serves as an unbiased community prevalence estimator. This is especially true with Covid-19 whose asymptomatic period is about a fortnight. Ultimately, WBE allows near real-time cheap monitoring of health indicators such as obesity, diabetes, drug use, microbial antibiotic resistance, and disease outbreak. Its use in disease outbreaks offers particularly rich data on genetic diversity of outbreaks and phylogenic analysis can reveal viral ancestry.
In Australia, the University of Queensland has been linking census data to wastewater samples across the country to see the interrelationship between wastewater chemicals and social and economic measures of a population. Doing that opened the study of socioeconomic influences on chemical consumption. This study showed that caffeine consumption is associated with aspects of financial capability and educational attainment in Australia for instance.
WBE success
WBE is successful in sentinel surveillance providing early outbreak warnings and in determining the efficacy of public health interventions. It is remarkably sensitive at picking up infections and viral load in wastewater. For polio for instance, WBE sensitivity is estimated at about 1 case per 10,000 uninfected people. WBE also allows spatial sensitivity by moving “upstream.” WBE can detect variations in circulating strains through phylogenic analysis allowing for comparisons between region and viral genomic evolution. Another important benefit of WBE is that it enables disease prevalence gauging by circumventing individual stigmatization which can arrive from clinical diagnosis (early AIDS research for instance).
SARS-Cov-2 Simplified WBE Procedure
In general all WBE follows the same process: pretreatment, concentration, recovery, secondary concentration, then detection. Detection normally means either molecular analysis or traditional culturing. In an International Water Association (IWA) webinar on 19 June 2020 Charles Gerba, an environmental microbiologist at the Water, Energy, and Sustainable Technology Center (WEST) in the University of Arizona provided an outline of how they were testing:
- Gather a 500 mL to 1 L sample of wastewater (grab or composite was not specified)
- Take a 100-250 mL aliquot to process
- Spike some samples with 229E to test efficiency
- Store at -80°C for future analysis
- Centrifuge to remove solids because some virus are lost to solids – in general about 100 mL would spin down to 1-3 mL
- RT-qPCR: biomarkers (gene targets) N1, N2, N3, E229. Normally N2 and E229 are used to ensure the signal is specific enough. N1 and N3 are typically dropped
Difficulties in WBE Interpretation
WBE sounds amazing and it truly is. It has already been used successfully to track public health threats from polio to alcohol and all these achievements for a field under 20 years old. Its full potential isn’t even near realized at this point. However, there are several issues in the field. The largest is the lack of standardization and inability to compare results between testing facilities. These two factors are intrinsically linked but one will not necessarily solve the other. Another set of issues revolve around tying total loads to population numbers.
Difficulties with standardization
WBE is still a new field. It has not decided upon standards for many common procedures yet. For instance, some areas preform pre-process techniques to lower the risk of catching Covid-19 from working with SARS-Cov-2. Different pre-processing techniques such as pasteurization or filtration, will produce different signal drops.
Even the sample collection is very different. In wastewater there are typically two kinds of sampling: grab and composite. Grab sampling reflects a discrete point in time and space; composite sampling essentially is several grab samples pooled together at regular time or spatial intervals. Composite sampling is the most common in wastewater because varying flow patterns cause hydraulic surges followed by intermittent periods of low to no flow. However, that does not necessarily make it the best method for WBE.
The solids amount in the wastewater can also reduce efficacy of RT-qPCR methods; what phase to analyze (particulate or liquid) can affect results. Likewise, different inhibitors used for sample shipment may reduce the signal strength. The specific method chosen as a standard unfortunately must consider cost as well as effectiveness and test time. Likewise decontamination procedures between tests must be considered.
Difficulties with linking viral loads to population cases
Sewers undergo infiltration and inflow (i/i). Infiltration is where groundwater enters the sewer system through joints or breaks, inflow is where water is channeled into a sewer from various sources into the sewer such as downspouts. Without getting too deep, there are combined, separate, and merged sewer systems referring to surface runoff or sewage removal. Most large cities have merged systems were sewers were initially built as combined but started providing separate runoff and sewage systems. In short, a remote lab won’t necessarily have the proper infrastructural or weather contextualization to interpret the RNA signal in testing.
Another significant hurdle for disease monitoring is figuring out each disease’s excretion pattern. While it may seem reasonable that a greater number of sick people or sicker people excrete a higher viral load this is not always the case. Extrapolating the viral load to clinical cases becomes complicated. If the disease already has a well known viral shedding pattern and spread pattern with significant effort based, on where in the outbreak a disease is, you can get a correlation however it would be predicated upon many assumptions. For diseases with well defined correlations between degree of illness and viral shedding combined with disease transmission knowledge it is not possible to distinguish between one moderately sick person and two or more asymptomatic people with any degree of precision. With novel diseases only trend analysis is possible. Given the unknowns around viral shedding it becomes difficult to determine how the RNA signal drop corresponds with prevalence drops in the local community. It also becomes difficult to determine how strong the signal change needs to be to differentiate from statistical noise.
Correlating viral loads with clinically identified cases becomes even more challenging because of variable excretion rates during the infection, temporal delays, inconsistent spatial variability due to travel leading to use of multiple wastewater treatment systems, i/i, inactivation during transport, or infrequent, absent or inadequate clinical testing. Genomic instability in wastewater, sampling variability (grab/composite), and viral concentration efficiency differences compound these problems.
Where the sample was taken from, for instance from the sewage network or treatment plant, is also believed to effect viral recovery making comparisons difficult. The type of upstream user, for example domestic or industrial, will make a large difference as well. Areas with more septic systems then become harder to check. Likewise there is a divide between smaller more rural populations and larger cities; cities tend to create more normalization and may not necessarily be compared to their rural counterparts.
Practical difficulties with WBE
The best monitoring schedule at what frequency and spatial resolutions remain open questions which most likely vary across diseases. Likewise, who pays for the monitoring is an important consideration. Currently, WEST’s price list is between $350 and $1,250 per sample depending on how exactly they perform and analyze the sample. The quantification level can be tricky as well since most PCR techniques were developed for the clinical setting instead of an environmental one. There is also a privacy issue with this sort of monitoring.
Conclusions
WBE is an amazing tool for disease monitoring but is better suited to looking at trends because direct comparisons across catchments remains elusive. Since some aspects rely on data individual to specific catchments (recent precipitation, sewer condition, length of sewer and viral decay in sewer transport etc…) direct comparisons between viral loads may never really be achieved.
Further Resources
US EPA on Coronavirus in water and wastewater
Research Centers:
- The WEST Center at the University of Arizona, the United States
- KWR Water Research Institute in Nieuwegein, the Netherlands
- The Queensland Alliance for Environmental Health Sciences at the University of Queensland, Australia
Papers:
- Kitajima, M., Ahmed, W., Bibby, K., Carducci, A., Gerba, C. P., Hamilton, K. A., … & Rose, J. B. (2020). SARS-CoV-2 in wastewater: State of the knowledge and research needs. Science of The Total Environment, 139076
- Nemudryi, A., Nemudraia, A., Surya, K., Wiegand, T., Buyukyoruk, M., Wilkinson, R., & Wiedenheft, B. (2020). Temporal detection and phylogenetic assessment of SARS-CoV-2 in municipal wastewater. medRxiv : the preprint server for health sciences, 2020.04.15.20066746
- Venugopal, Anila, Harsha Ganesan, Suresh Selvapuram Sudalaimuthu Raja, Vivekanandhan Govindasamy, Manimekalan Arunachalam, Arul Narayanasamy, Palanisamy Sivaprakash et al. “Novel Wastewater Surveillance Strategy for Early Detection of COVID–19 Hotspots.” Current Opinion in Environmental Science & Health (2020)
- Ahmed, W., Angel, N., Edson, J., Bibby, K., Bivins, A., O’Brien, J. W., … & Tscharke, B. (2020). First confirmed detection of SARS-CoV-2 in untreated wastewater in Australia: A proof of concept for the wastewater surveillance of COVID-19 in the community. Science of The Total Environment, 138764
- Gracia-Lor, E., Castiglioni, S., Bade, R., Been, F., Castrignanò, E., Covaci, A., … & Lai, F. Y. (2017). Measuring biomarkers in wastewater as a new source of epidemiological information: Current state and future perspectives. Environment international, 99, 131-150
- Xagoraraki, I., & O’Brien, E. (2020). Wastewater-based epidemiology for early detection of viral outbreaks. In Women in Water Quality (pp. 75-97). Springer, Cham